- A machine learning approach developed by researchers at MIT’s Koch Institute and Massachusetts General Hospital (MGH) may aid in cancer diagnosis of the unknown primary by examining gene expression programs associated with early cell development and differentiation.
- The scientists focused the model on indicators of disrupted developmental pathways in cancer cells to find a compromise between lowering the number of characteristics while still capturing the most essential information.
- The researchers subsequently created the Developmental Multilayer Perceptron (D-MLP), a machine-learning model that rates a tumor for its developmental components and forecasts its origin.
- After training, the D-MLP was applied to 52 fresh samples of especially difficult malignancies of unknown origin that could not be classified using existing techniques.
- Furthermore, comprehensive comparisons of tumor and embryonic cells in the study offered promising and sometimes surprising insights into the gene expression patterns of different tumor types.
The first stage in deciding the best therapy for a cancer patient is identifying their exact form of cancer, which includes pinpointing the main site, the organ or portion of the body where the disease develops.
Even with rigorous testing, the cause of cancer cannot always be established. Although these cancers of unclear origin are often aggressive, oncologists must treat them with non-targeted medicines, which typically have severe side effects and result in low survival rates.
Using machine learning for cancer diagnosis
A new machine learning methodology developed by researchers at MIT’s Koch Institute for Integrative Cancer Research and Massachusetts General Hospital (MGH) may aid in classifying cancers of the unknown primary by examining gene expression programs associated with early cell development and differentiation.
Salil Garg, a pathologist at MGH and a Charles W. (1955) and Jennifer C. Johnson Clinical Investigator at the Koch Institute, stated: “Sometimes you can apply all the tools that pathologists have to offer, and you are still left without an answer. Machine learning tools like this one could empower oncologists to choose more effective treatments and give more guidance to their patients.”
Garg is the senior author of new research published on August 30 in Cancer Discovery, and the main author is MIT postdoc Enrico Moiso. The artificial intelligence technique has high sensitivity and accuracy in recognizing cancer types.
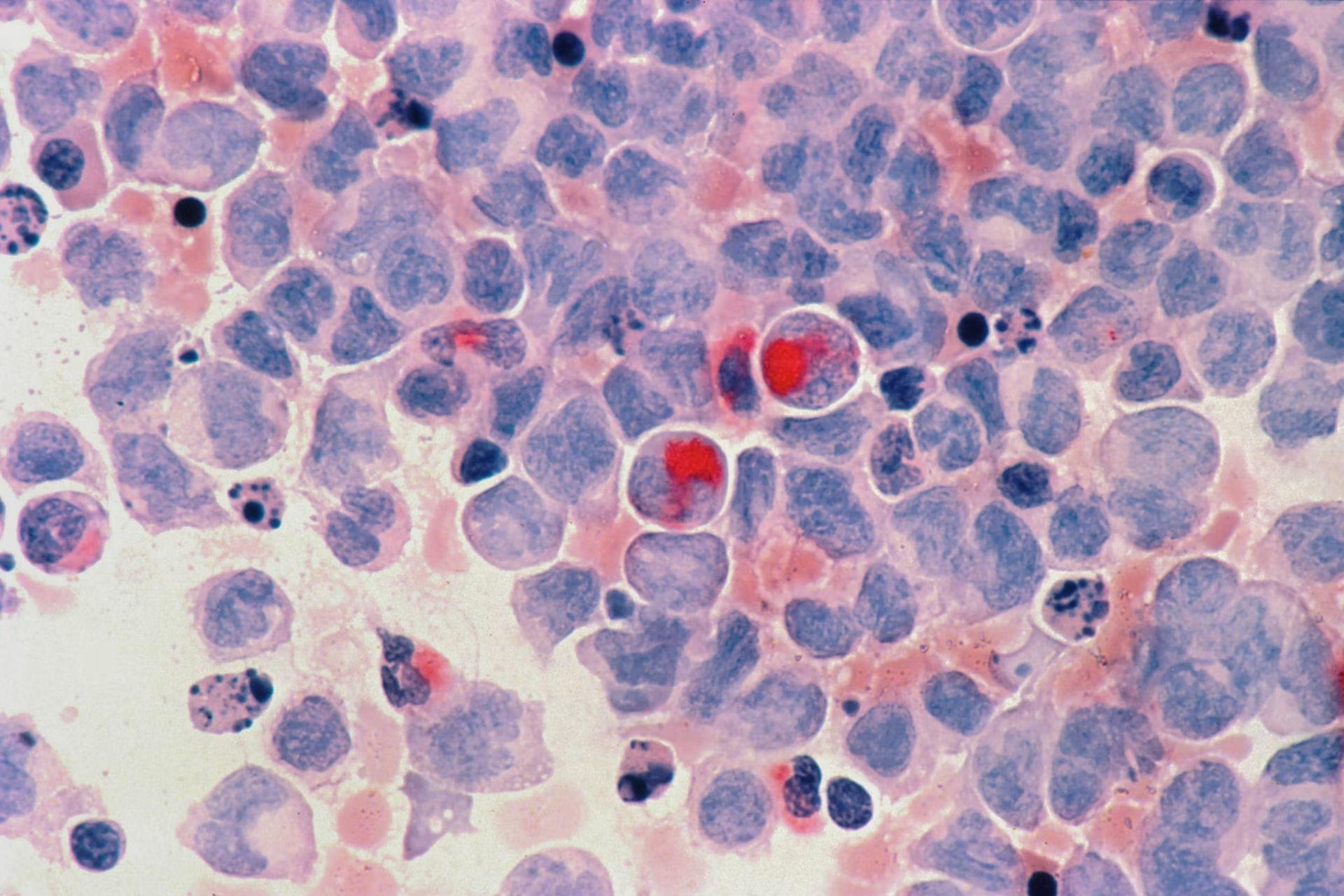
Parsing the changes in gene expression across various cancers from an unknown source is a great challenge for machine learning to handle. Cancer cells appear and function quite differently than normal cells, due in part to substantial changes in how their genes are expressed.
Advances in single-cell profiling and efforts to classify distinct cell expression patterns in cell atlases have resulted in a plethora of, though sometimes overwhelming, data, including clues to how and where different malignancies started.
Building a machine learning model that utilizes distinctions between healthy and normal cells, as well as between different types of cancer, into a diagnostic tool, on the other hand, is a balancing act. Suppose a very sophisticated model accounts for too many aspects of cancer gene expression. In that case, it may appear to learn the training data flawlessly yet stumble when confronted with fresh data.
However, by simplifying the model by reducing the number of characteristics, the model may lose information that might lead to correct cancer classifications.
Neural network-based visual stimuli classification paves the way for early Alzheimer’s diagnosis
The scientists focused the model on indicators of disrupted developmental pathways in cancer cells to find a compromise between lowering the number of characteristics while still capturing the most essential information. Many pathways direct how cells reproduce, expand, change their form, and move as an embryo grows, as undifferentiated cells specialize into distinct organs.
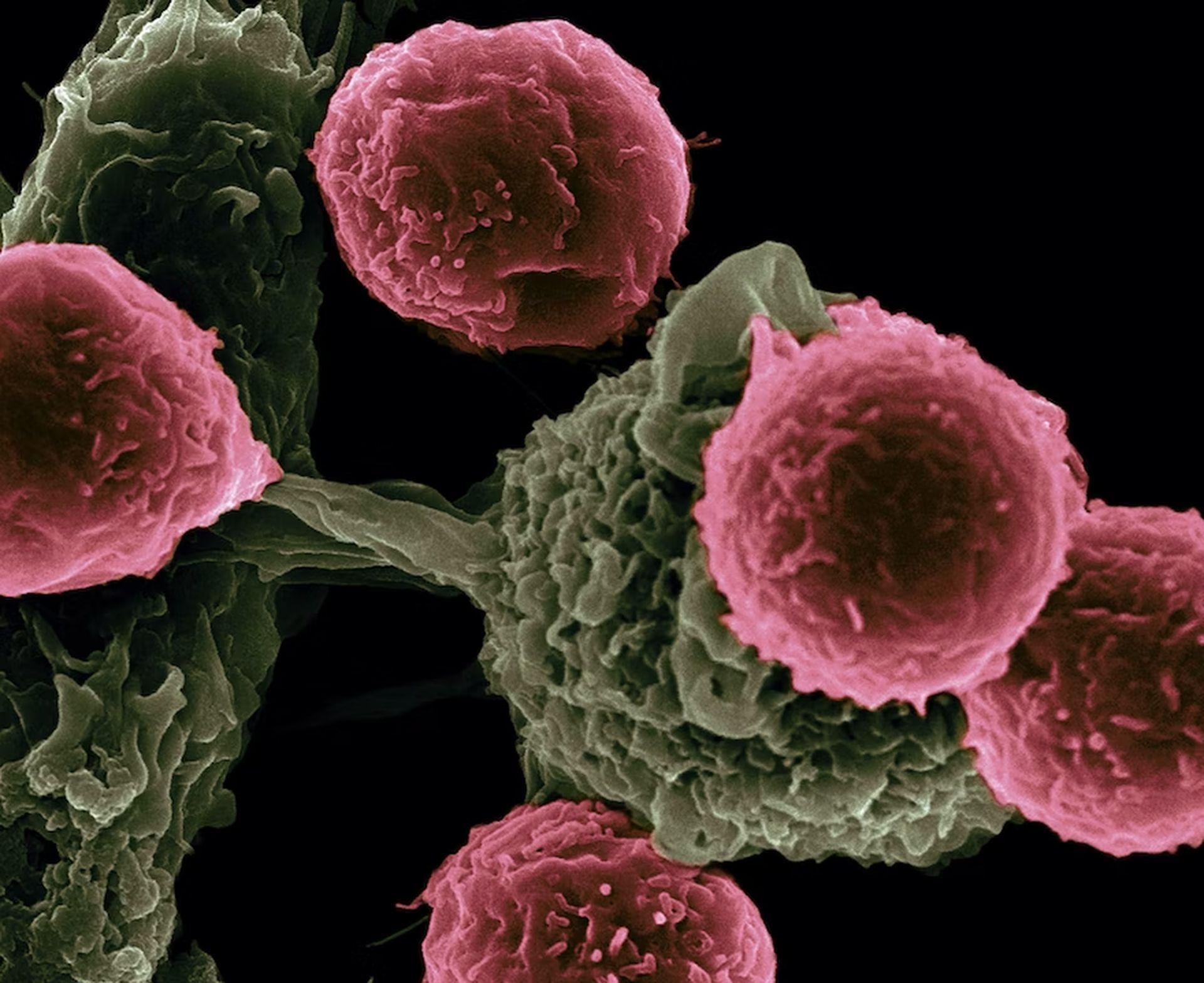
Cancer cells lose many of their specific characteristics as the tumor grows. At the same time, as they obtain the ability to multiply, change, and metastasize to other tissues, they begin to resemble embryonic cells in certain aspects. Many gene expression programs that control embryogenesis are reactivated or dysregulated in cancer cells.
Creating an ML algorithm that can diagnose cancer
The researchers contrasted the Cancer Genome Atlas (TCGA), which provides gene expression data for 33 tumor types, with the Mouse Organogenesis Cell Atlas (MOCA), which examines 56 distinct trajectories of embryonic cells as they grow and differentiate.
Moiso explains, “Single-cell resolution tools have dramatically changed how we study the biology of cancer, but how we make this revolution impactful for patients is another question. With the emergence of developmental cell atlases, especially ones that focus on early phases of organogenesis such as MOCA, we can expand our tools beyond histological and genomic information and open doors to new ways of profiling and identifying tumors and developing new treatments.”
Researchers found a new ML method capable of counting cells for disease diagnosis
The map of correlations between developmental gene expression patterns in tumors and embryonic cells was then used to train a machine learning algorithm. The researchers divided the gene expression of TCGA tumor samples into discrete components corresponding to a certain moment in a developmental trajectory and assigned a mathematical value to each component. The researchers subsequently created the Developmental Multilayer Perceptron (D-MLP), a machine-learning model that rates a tumor for its developmental components and forecasts its origin.
Following training, the D-MLP was applied to 52 fresh samples of especially difficult malignancies of unknown origin that could not be identified using existing methods. These were the most difficult patients seen at MGH in a four-year period commencing in 2017. Excitingly, the model classified the tumors into four groups and produced forecasts and other data that might aid in diagnosing and treating these patients.
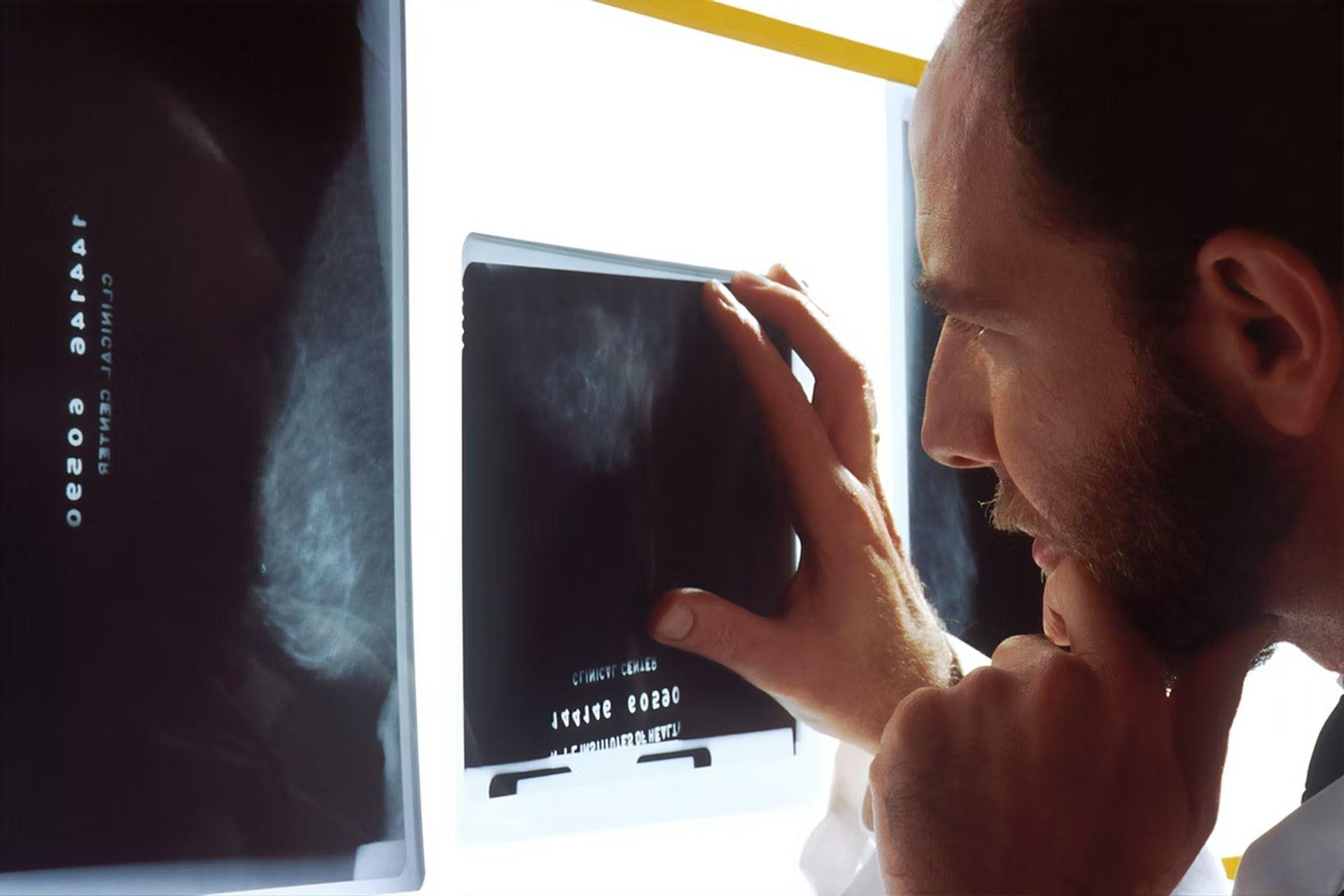
One sample, for example, came from a woman who had a history of breast cancer and had evidence of an aggressive tumor in the fluid spaces surrounding the abdomen. Using the available methods, oncologists could not locate a tumor mass or categorize cancer cells. However, D-MLP strongly predicted ovarian cancer. Six months after the patient originally came, a lump in the ovary was discovered to be the source of the malignancy.
Furthermore, the study’s comprehensive comparisons of tumor and embryonic cells offered promising and sometimes surprising insights into the gene expression profiles of different tumor types. For example, during the early stages of embryonic development, a rudimentary gut tube emerges, with the foregut producing the lungs and other adjacent organs and the mid-and hindgut constituting much of the digestive tract.
Machine learning makes life easier for data scientists
The study found that lung-derived tumor cells had substantial parallels not just to the foregut but also to the mid-and hindgut-derived developmental trajectories. These findings imply that variations in developmental programs may one day be used in the same manner that genetic mutations are frequently used to generate tailored or targeted cancer therapies.
While the work provides a robust technique for tumor classification, it does have certain drawbacks. In the future, researchers intend to improve the prediction value of their model by including more forms of data, namely information from radiography, microscopy, and other types of tumor imaging.
Garg stated: “Developmental gene expression represents only one small slice of all the factors that could be used to diagnose and treat cancers. Integrating radiology, pathology, and gene expression information together is the true next step in personalized medicine for cancer patients.”






